Table of Contents
Protein Groups and peptides
See also how-to access the protein group view and/or how-to access peptide views.
Definition
A Protein Group (similar to Mascot® protein hit) is a collection of proteins identified by a set of peptides. A Protein Group contains not only all proteins covered by the same set of peptides but also all proteins covered by a subset of these peptides. In details, a Protein Group:
- is represented by a master (or typical) protein
- contains a collection of peptides identifying this master protein
- contains a collection of proteins having the sameset of peptides
- contains a collection of proteins having a subset of peptides
A Protein Group is always attached to a Context, Search or User Context. For the latter, protein groups are obtained when running the Protein Grouping algorithm on a given context.
Mode details about protein group tree view
Protein groups are associated to a context, thus it is possible to browse protein groups for a specific context.
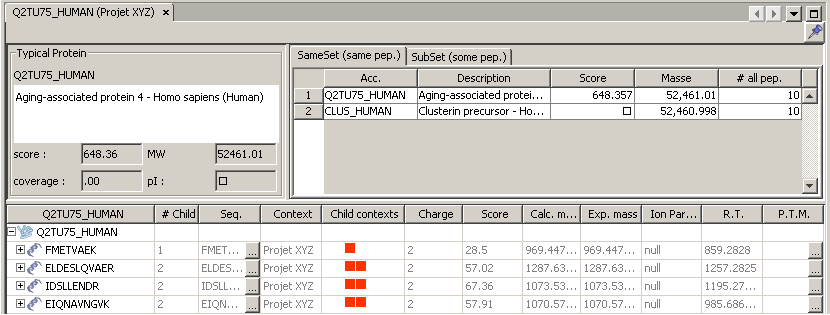
 This window list all protein groups by their typical protein accession (represented by
This window list all protein groups by their typical protein accession (represented by  ). At least, the following properties are displayed :
). At least, the following properties are displayed :
typicalprotein accession- typical protein
score - number of
peptides - number of
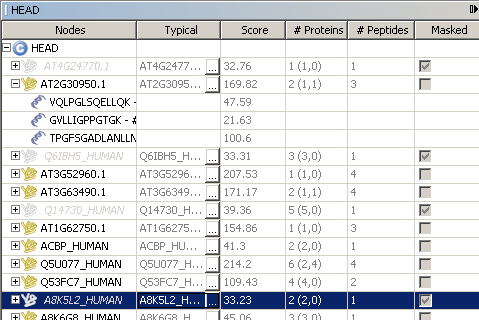
proteinsin group (in sameset, in subset). maskedspecify if protein group is masked (after a filtereing )
If other properties are associated to the protein group or typical protein, such as spectral count, they could also be displayed (see properties general description page)
For each group a list of all peptides used to identify the proteins is also displayed.
Masked protein groups are displayed in gray and italic.
Mode details about protein group details view
hEIDI allow users to access protein group's detail from different place.
The protein group window shows
- details about this typical protein
- lists of group content: sameset/subset proteins and peptides. Masked proteins are displayed in gray and italic.
If you select another protein from the sameset/subset tabs, the table showing peptides will be updated accordingly. Only peptide matching the selected protein will be displayed.
More details about peptides view
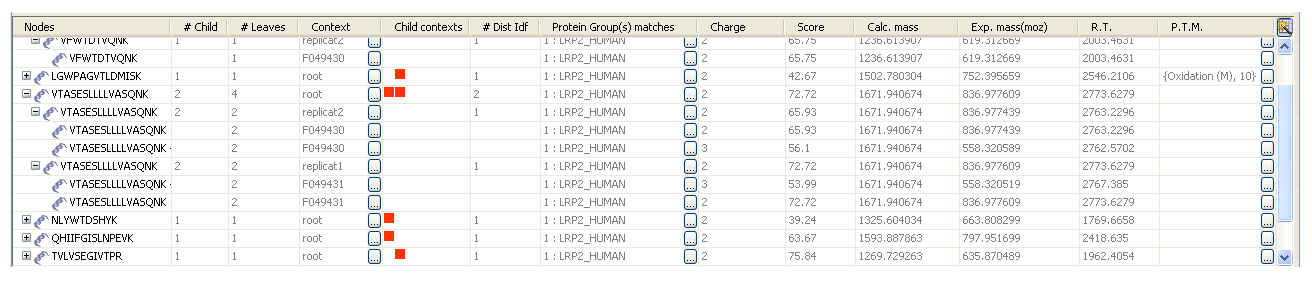
Peptide information is visible in the protein group view (context's protein group list or protein group details, see above) but also in the list of all peptides of a context.
A '+' appears in front of each peptide node. If you deploy it, you will explore the child peptide, i.e. the peptides that belong to the contexts below
- # Child: number of direct child peptides, i.e. the number of peptides in the context(s) directly below
- # Leaves: number of spectra
- Context: context name
- Child contexts: for each peptide node, small coloured squares correspond to the context(s) to which belong to the child peptides. The colour changes according the depth of the context. When positioning the mouse cursor onto this image, the context names are displayed as a tooltip. The context of the last child peptide is not represented (a context is actually an identification result in this case).
- # Dist Idf: number of distinct identifications from which child peptides belong to
- Protein Group matche(s): number of protein match(es) and concerned typical protein(s)
- Charge: charge of the best child
- Score: score of the best child
- Calc. mass: calculated mass
- Exp. mass: experimental mass of the best child
- R.T.: retention time of the best child
- P.T.M.: post-traductional modifications