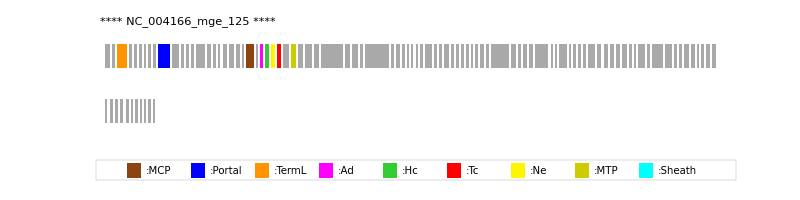
| Protein Superfamily | Index in Query Genome | Sequence Header | Detection Method |
|---|---|---|---|
| MCP | 23 | lcl|Aclame:protein:vir:5974|NCBI_annot:hypothetical | HHsearch (proba=100.00%) |
| Portal | 10 | lcl|Aclame:protein:vir:5961|NCBI_annot:hypothetical | HHsearch (proba=100.00%) |
| TermL | 3 | lcl|Aclame:protein:vir:5954|NCBI_annot:hypothetical | Blast (e-value=0) |
| MTP | 30 | lcl|Aclame:protein:vir:5980|NCBI_annot:hypothetical | Blast (e-value=1e-101) |
| Sheath | NOT FOUND | ||
| Ad1 | 25 | lcl|Aclame:protein:vir:5976|NCBI_annot:hypothetical | HHsearch (proba=100.00%) |
| Hc1 | 26 | lcl|Aclame:protein:vir:5977|NCBI_annot:hypothetical | HHsearch (proba=100.00%) |
| Ne1 | 27 | lcl|Aclame:protein:vir:5978|NCBI_annot:hypothetical | HHsearch (proba=100.00%) |
| Tc1 | 28 | lcl|Aclame:protein:vir:5979|NCBI_annot:hypothetical | HHsearch (proba=100.00%) |
| Ad2 | NOT FOUND | ||
| Hc2 | NOT FOUND | ||
| Tc2 | NOT FOUND | ||
| Ad3 | NOT FOUND | ||
| Hc3 | NOT FOUND | ||
| Ad4 | NOT FOUND |
| Protein Superfamily | Corresponding Protein in Aclame | Aclame Phage | Seq. Identity (%) |
|---|---|---|---|
| MCP | protein:vir:5974 protein:vir:102944 protein:vir:94989 | SPP1 EJ-1 KS7 | 100 % 41 % 34 % |
| Portal | protein:vir:5961 protein:vir:79043 protein:vir:97336 | SPP1 phiC2 52A | 100 % 36 % 34 % |
| TermL | protein:vir:5954 protein:vir:9870 protein:vir:105297 | SPP1 315.5 PH15 | 100 % 62 % 60 % |
| MTP | protein:vir:5980 protein:vir:94505 protein:vir:9932 | SPP1 phiJL-1 315.6 | 100 % 39 % 35 % |
| Ad1 | protein:vir:5976 protein:vir:97430 protein:vir:95071 | SPP1 92 X2 | 100 % 37 % 37 % |
| Hc1 | protein:vir:5977 protein:vir:1890 protein:vir:96294 | SPP1 HK022 ROSA | 100 % 27 % 27 % |
| Ne1 | protein:vir:5978 protein:vir:97327 protein:vir:1243 | SPP1 52A phi ETA | 100 % 43 % 43 % |
| Tc1 | protein:vir:5979 protein:vir:105337 protein:vir:97421 | SPP1 PH15 92 | 100 % 30 % 30 % |