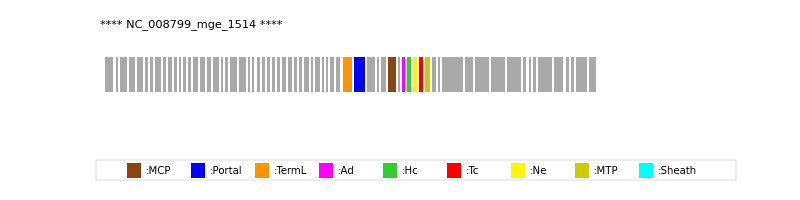
| Protein Superfamily | Index in Query Genome | Sequence Header | Detection Method |
|---|---|---|---|
| MCP | 46 | lcl|Aclame:protein:vir:105905|NCBI_annot:major | HHsearch (proba=100.00%) |
| Portal | 42 | lcl|Aclame:protein:vir:105889|NCBI_annot:portal | HHsearch (proba=100.00%) |
| TermL | 41 | lcl|Aclame:protein:vir:105897|NCBI_annot:large | Blast (e-value=0) |
| MTP | 52 | lcl|Aclame:protein:vir:105912|NCBI_annot:major | Blast (e-value=1e-108) |
| Sheath | NOT FOUND | ||
| Ad1 | 48 | lcl|Aclame:protein:vir:105899|NCBI_annot:head | HHsearch (proba=100.00%) |
| Hc1 | 49 | lcl|Aclame:protein:vir:105908|NCBI_annot:hypothetical | HHsearch (proba=100.00%) |
| Ne1 | 50 | lcl|Aclame:protein:vir:105916|NCBI_annot:hypothetical | HHsearch (proba=100.00%) |
| Tc1 | 51 | lcl|Aclame:protein:vir:105892|NCBI_annot:tail | HHsearch (proba=100.00%) |
| Ad2 | NOT FOUND | ||
| Hc2 | NOT FOUND | ||
| Tc2 | NOT FOUND | ||
| Ad3 | NOT FOUND | ||
| Hc3 | NOT FOUND | ||
| Ad4 | NOT FOUND |
| Protein Superfamily | Corresponding Protein in Aclame | Aclame Phage | Seq. Identity (%) |
|---|---|---|---|
| MCP | protein:vir:105905 protein:vir:94142 protein:vir:41 | phiETA3 96 A118 | 100 % 100 % 44 % |
| Portal | protein:vir:105889 protein:vir:94101 protein:vir:9871 | phiETA3 96 315.5 | 100 % 100 % 26 % |
| TermL | protein:vir:105897 protein:vir:94084 protein:vir:4897 | phiETA3 96 Sfi11 | 100 % 100 % 51 % |
| MTP | protein:vir:96311 protein:vir:105912 protein:vir:94128 | ROSA phiETA3 96 | 100 % 100 % 100 % |
| Ad1 | protein:vir:105899 protein:vir:94126 protein:vir:96831 | phiETA3 96 EW | 100 % 100 % 31 % |
| Hc1 | protein:vir:96294 protein:vir:105908 protein:vir:94092 | ROSA phiETA3 96 | 100 % 100 % 97 % |
| Ne1 | protein:vir:105916 protein:vir:94108 protein:vir:96288 | phiETA3 96 ROSA | 100 % 99 % 89 % |
| Tc1 | protein:vir:96260 protein:vir:105892 protein:vir:94096 | ROSA phiETA3 96 | 100 % 100 % 100 % |