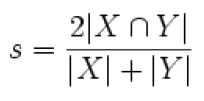userguide:contextdefinition
This is an old revision of the document!
Table of Contents
Context comparisons
Definition
The purpose is to search for similar protein groups, i.e. protein groups:
- having the same set of proteins
- identified by a same set of peptides
The Dice coefficient is used to measure similarity (result ranges from 0 to 1).
Compare two contexts with each other
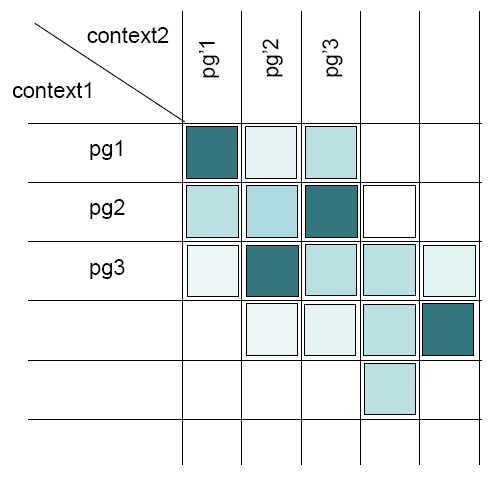 It compares every protein groups of the first context with every protein groups of the second context.
It compares every protein groups of the first context with every protein groups of the second context.
Comparing two protein groups each other means:
- Checking the similarity of their protein sets (
 sameset and subset)
sameset and subset) - Checking the similarity of their peptide sets
- Checking if “master” proteins are identical
Compare N contexts
This comparison is similar to the previous one: similarity measure and  “best alignment”
“best alignment”
Each context is compared with a “unique reference”. First of all, this reference is built from the identification union, i.e. the parent context.
GUI: Compare contexts each other
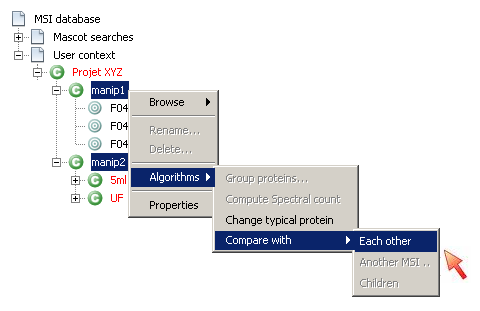
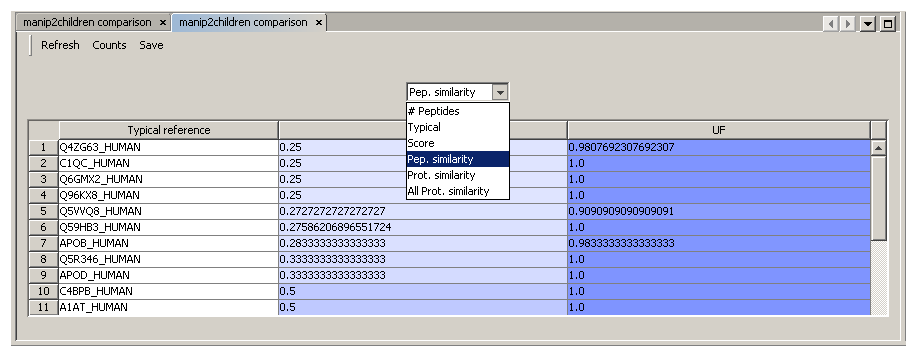
- Select the two contexts (for which protein grouping has been executed) you want to compare each other
- Right-click and select “Algorithms→Compare with→Each Other”
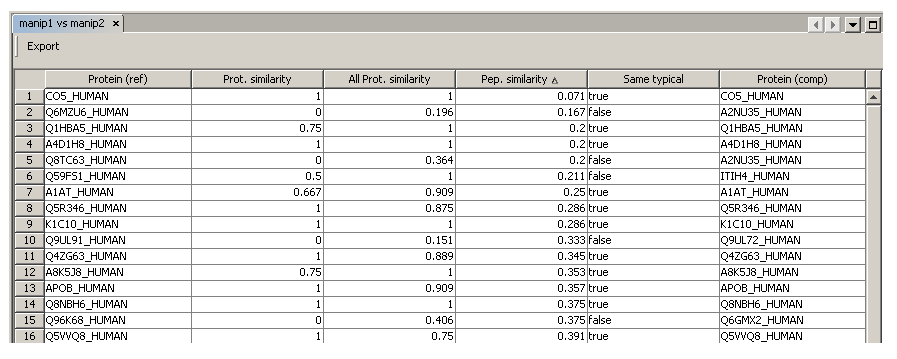
- The comparison window display the result
- You can save results as a .xls, .pdf or .html file using the “Export” button
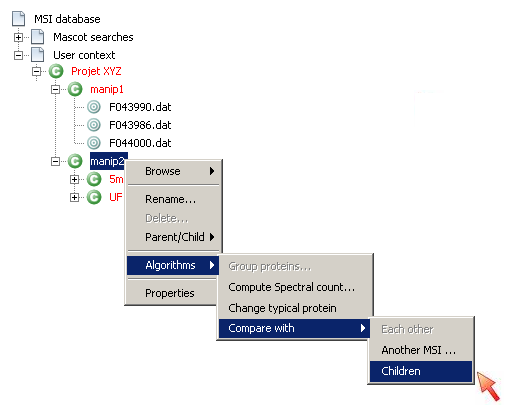
GUI: Compare context with its children
userguide/contextdefinition.1260950987.txt.gz · Last modified: 2009/12/16 09:09 by 132.168.74.230