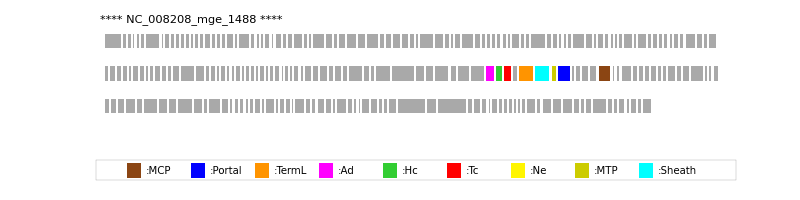
| Protein Superfamily | Index in Query Genome | Sequence Header | Detection Method |
|---|---|---|---|
| MCP | 156 | lcl|Aclame:protein:vir:100603|NCBI_annot:gp23 | HHsearch (proba=100.00%) |
| Portal | 151 | lcl|Aclame:protein:vir:100598|NCBI_annot:gp20 | HHsearch (proba=100.00%) |
| TermL | 148 | lcl|Aclame:protein:vir:100538|NCBI_annot:gp17 | Blast (e-value=0) |
| MTP | 150 | lcl|Aclame:protein:vir:100597|NCBI_annot:gp19 | Blast (e-value=2e-94) |
| Sheath | 149 | lcl|Aclame:protein:vir:100539|NCBI_annot:gp18 | HHsearch (proba=100.00%) |
| Ad1 | NOT FOUND | ||
| Hc1 | NOT FOUND | ||
| Ne1 | NOT FOUND | ||
| Tc1 | NOT FOUND | ||
| Ad2 | 144 | lcl|Aclame:protein:vir:100534|NCBI_annot:gp13 | HHsearch (proba=100.00%) |
| Hc2 | 145 | lcl|Aclame:protein:vir:100535|NCBI_annot:gp14 | HHsearch (proba=100.00%) |
| Tc2 | 146 | lcl|Aclame:protein:vir:100536|NCBI_annot:gp15 | HHsearch (proba=100.00%) |
| Ad3 | NOT FOUND | ||
| Hc3 | NOT FOUND | ||
| Ad4 | NOT FOUND |
| Protein Superfamily | Corresponding Protein in Aclame | Aclame Phage | Seq. Identity (%) |
|---|---|---|---|
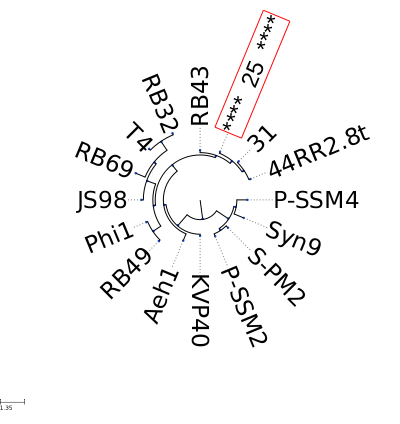
| MCP | protein:vir:100603 protein:vir:101039 protein:vir:101811 | 25 44RR2.8t 31 | 100 % 93 % 92 % |
| Portal | protein:vir:100598 protein:vir:101189 protein:vir:101806 | 25 44RR2.8t 31 | 100 % 91 % 91 % |
| TermL | protein:vir:100538 protein:vir:101186 protein:vir:101803 | 25 44RR2.8t 31 | 100 % 87 % 86 % |
| MTP | protein:vir:100597 protein:vir:101188 protein:vir:101805 | 25 44RR2.8t 31 | 100 % 88 % 88 % |
| Sheath | protein:vir:100539 protein:vir:101187 protein:vir:101804 | 25 44RR2.8t 31 | 100 % 86 % 86 % |
| Ad2 | protein:vir:100534 protein:vir:101153 protein:vir:101857 | 25 44RR2.8t 31 | 100 % 84 % 84 % |
| Hc2 | protein:vir:100535 protein:vir:101154 protein:vir:101800 | 25 44RR2.8t 31 | 100 % 82 % 82 % |
| Tc2 | protein:vir:100536 protein:vir:101155 protein:vir:101801 | 25 44RR2.8t 31 | 100 % 80 % 80 % |