userguide:spectralcount
This is an old revision of the document!
Table of Contents
Spectral count
Definition
The Spectral Count algorithm can be run for a given context and will loop on every protein groups of this context to calculate this property.
Spectral count reflects relative abundance of a protein by counting how many times all peptides of a proteins have been fractioned.
- For Mascot context the spectral count is similar to the number of peptides (which return number of all peptides : similar and duplicate).
- For user context, this algorithm will count how many leaf peptides have been grouped for each peptide of the protein group, thus spectral count and number of peptides are different.
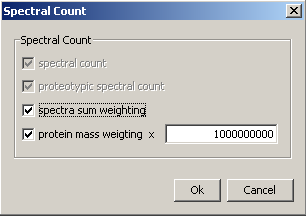
Variant values can also be calculated :
- Spectra sum weighting : divide all spectral count values by the number of spectra in the context
- Protein mass weighting : divide all spectral count values by each master protein mass; A Scale factor can be applied to all Protein mass weighting spectral count values
 Several other characteristics may be useful to collect.
Several other characteristics may be useful to collect.
Run Spectral Count algorithm
- Select the context of your choice (for which protein grouping have been executed)
- Right-click and select
Algorithms > Compute Spectral Count - Spectral count results are available in Protein Group
userguide/spectralcount.1271248043.txt.gz · Last modified: 2010/04/14 14:27 by 132.168.73.247