how_to:generateamtdb
This is an old revision of the document!
Table of Contents
Generate a new AMT database from a context
To generate a new AMT database in the MS Access format (.mdf file) from a given User context, follow these steps:
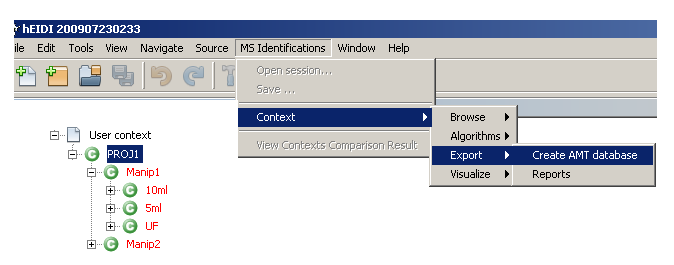
- Select the User context you want to export
- Select
MS Identifications > Context > Export > Create AMT databasefrom the main menu bar. - Browse to a folder where you want to save the AMT database file and click
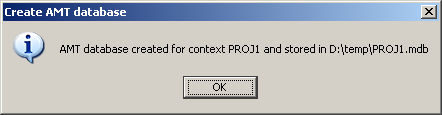
Open. - A confirmation dialog will indicate when saving process is finished.
Note : Before generating a new AMT db from a context, protein grouping should have been executed !
AMT database filters & Details about some field calculations
How specific values are calculated:
- MonoistopicMass: calculated_mass
- NET: (Retention Time/60)-5)/80
- PNET : null
- MS_MS_Obs_Count: # of child species
- sequence: species sequence
- High_Normalized_Score: score (max of child species scores)
- High_Discriminant_Score: null
When exporting an AMTdb, the following filters are applied (suppression):
- Reverse proteins
- Species with a sequence < 7
- Proteins identified by only 1 species with a score < 60
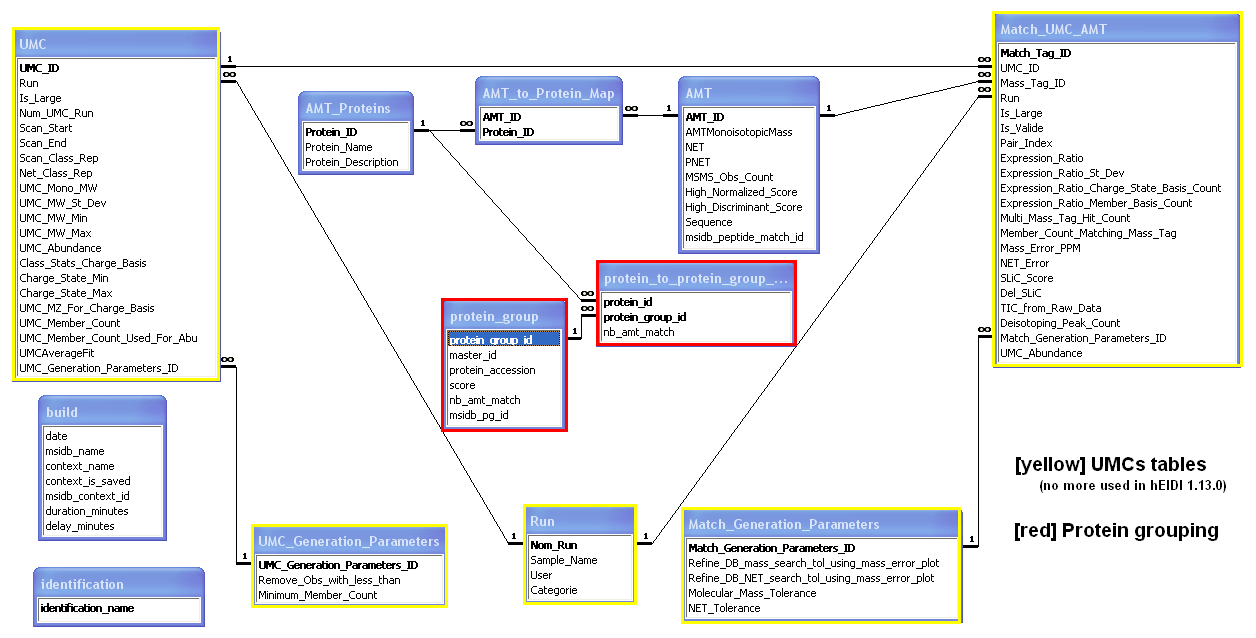
AMT database model
Below is the actual AMT database model showing relationships between tables in MS Access.
When exporting an AMT database from hEIDI, 5 tables are populated:
- Table AMT: species of the exported User context
- Table AMT_Proteins: proteins on which species match
- Protein_ID: protein identifier
- Protein_name: protein accession
- Table AMT_to_Protein_Map: map table between AMT & proteins
- Table protein_group: protein groups in which proteins are gathered
- protein_group_id: protein group identifier
- master_id: master protein identifier
- protein_accession: master protein accession
- score: master protein score
- nb_amt_match: # of peptides that match on master protein
- msidb_pg_id: protein group identifier in MSIdb (protein_group table)
- Table protein_to_protein_group_map: map table between proteins & protein groups
- protein_id: protein identifier
- protein_group_id: protein group identifier
- nb_amt_match: # of peptides that match on this protein in this protein group
- Table identification: identifications used in the exported User context
- identification_name: names of identifications included in the exported User context
- Table build: general information
- date: AMTdb creation date
- msidb_name: MSIdb name
- context_name: name of the exported context
- context_is_saved: is the exported context has been saved to MSIdb ?
- msidb_context_id: context identifier in the MSIdb (look at this field only if the context has been saved to MSIdb)
UMCs tables are populated when using 'Read UMCs' tool.
how_to/generateamtdb.1280822906.txt.gz · Last modified: 2010/08/03 10:08 by 132.168.74.230