- PNET : null
- MS_MS_Obs_Count: count(peptide_id) i.e. # of grouped species
- High_Normalized_Score: max(score(peptide))
- Reverse proteins
- Species with a sequence < 7
- Proteins identified by only 1 species with a score < 60
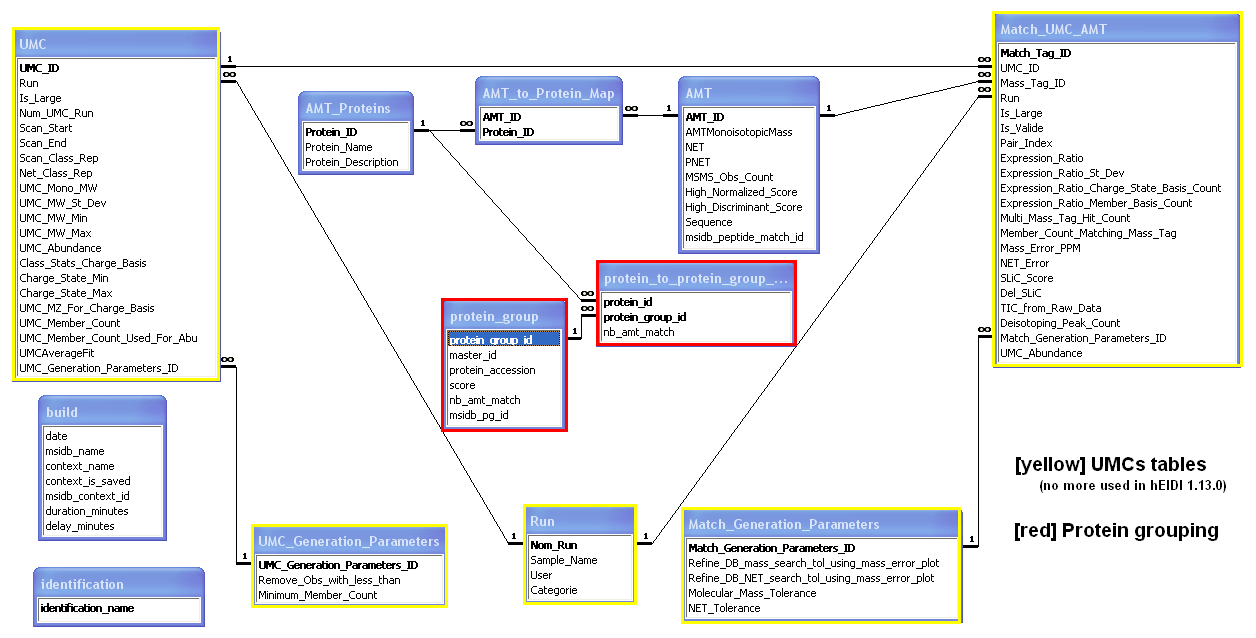
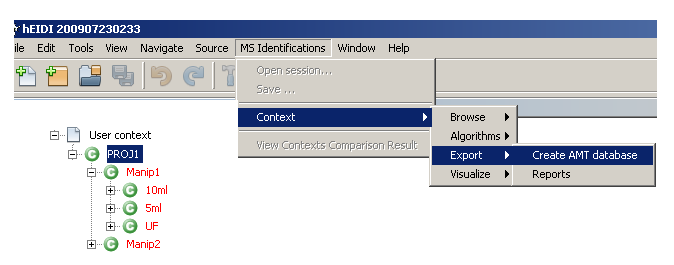
When exporting an AMT database from hEIDI, 5 tables are populated:
- Table AMT: species of the exported User context
- Table AMT_Proteins: proteins on which species match
- Protein_ID: protein identifier
- Protein_name: protein accession
- Table AMT_to_Protein_Map: map table between AMT & proteins
- Table protein_group: protein groups in which proteins are gathered
- protein_group_id: protein group identifier
- master_id: master protein identifier
- protein_accession: master protein accession
- score: master protein score
- nb_amt_match: # of peptides that match on master protein
- msidb_pg_id: protein group identifier in MSIdb (protein_group table)
- Table protein_to_protein_group_map: map table between proteins & protein groups
- protein_id: protein identifier
- protein_group_id: protein group identifier
- nb_amt_match: # of peptides that match on this protein in this protein group
- Table identification: identifications used in the exported User context
- identification_name: names of identifications included in the exported User context
- Table build: general information
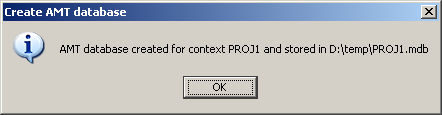
- date: AMTdb creation date
- msidb_name: MSIdb name
- context_name: name of the exported context
- context_is_saved: is the exported context has been saved to MSIdb ?
- msidb_context_id: context identifier in the MSIdb (look at this field only if the context has been saved to MSIdb)