how_to:accessadjustedspectralcountdata
Table of Contents
How to run and access adjusted spectral (ASC) count data
See spectralcount for more details on the algorithm and parameters.
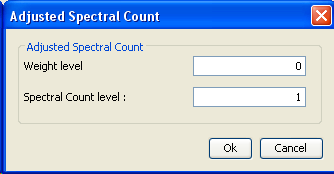
Run the ASC algorithm
- Select one or more context of your choice (for which protein grouping have been executed)
- Right-click and select
Algorithms > Compute Adjusted Spectral Count - Save the MSI in order to store properties.
View ASC data
Actually the ASC result is only accessible
- directly after ASC have been run : See Bellow
- in comparison result : view comparemulticontexts
after ASC have been run
In the output windows, (menu View > IDE Log), algorithm information is display between — STAT ASC Result Report — and — END ASC Result Report —. Copy and paste the output into Excel to view
- The first lines, gives the contexts involved in ASC.
- Then first part display lines formatted as bellow:
| accession | Replicate S1 | Replicate E1 | ||||||
| AT1G65260.1 | Replicate S1= | PROTEO:4 | TOTAL:4 | WEIGHTED:4.0 | Replicate E1= | PROTEO:38 | TOTAL:38 | WEIGHTED:38.0 |
| AT1G07320.3 | Replicate S1= | PROTEO:15 | TOTAL:15 | WEIGHTED:15.0 | Replicate E1= | PROTEO:3 | TOTAL:3 | WEIGHTED:3.0 |
| AT5G15530.1 | Replicate S1= | PROTEO:1 | TOTAL:2 | WEIGHTED:1.1764706 | Replicate E1= | PROTEO:0 | TOTAL:2 | WEIGHTED:0.33333334 |
| AT1G80380.3 | Replicate S1= | PROTEO:15 | TOTAL:15 | WEIGHTED:15.0 | Replicate E1= | PROTEO:4 | TOTAL:4 | WEIGHTED:4.0 |
| AT1G27300.1 | Replicate S1= | PROTEO:- | TOTAL:- | WEIGHTED:- | Replicate E1= | PROTEO:- | TOTAL:- | WEIGHTED:- |
- And the second part display following lines :
| accession | Replicate S1 | Replicate E1 |
| AT1G65260.1 | Replicate S1=ProteinValues{weight=0, proteoSC=4, totalSC=4, weightedSC=4.0} | Replicate E1=ProteinValues{weight=0, proteoSC=38, totalSC=38, weightedSC=38.0} |
| AT1G07320.3 | Replicate S1=ProteinValues{weight=0, proteoSC=15, totalSC=15, weightedSC=15.0} | Replicate E1=ProteinValues{weight=0, proteoSC=3, totalSC=3, weightedSC=3.0} |
| AT5G15530.1 | Replicate S1=ProteinValues{weight=0, proteoSC=1, totalSC=2, weightedSC=1.1764706} | Replicate E1=ProteinValues{weight=0, proteoSC=0, totalSC=2, weightedSC=0.33333334} |
how_to/accessadjustedspectralcountdata.txt · Last modified: 2013/01/28 15:31 by 132.168.72.131