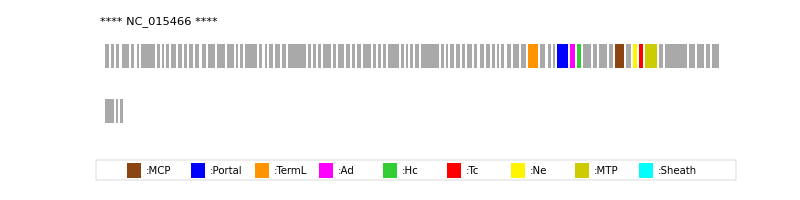
| Protein Superfamily | Index in Query Genome | Sequence Header | Detection Method |
|---|---|---|---|
| MCP | 74 | lcl|NC_015466.1_cdsid_YP_004421842.1 | HHsearch (proba=100.00%) |
| Portal | 67 | lcl|NC_015466.1_cdsid_YP_004421835.1 | HHsearch (proba=100.00%) |
| TermL | 63 | lcl|NC_015466.1_cdsid_YP_004421831.1 | Blast (e-value=6e-59) |
| MTP | 78 | lcl|NC_015466.1_cdsid_YP_004421846.1 | Blast (e-value=2e-56) |
| Sheath | NOT FOUND | ||
| Ad1 | 68 | lcl|NC_015466.1_cdsid_YP_004421836.1 | HHsearch (proba=100.00%) |
| Hc1 | 69 | lcl|NC_015466.1_cdsid_YP_004421837.1 | HHsearch (proba=97.39%) |
| Ne1 | 76 | lcl|NC_015466.1_cdsid_YP_004421844.1 | HHsearch (proba=100.00%) |
| Tc1 | 77 | lcl|NC_015466.1_cdsid_YP_004421845.1 | HHsearch (proba=100.00%) |
| Ad2 | NOT FOUND | ||
| Hc2 | NOT FOUND | ||
| Tc2 | NOT FOUND | ||
| Ad3 | NOT FOUND | ||
| Hc3 | NOT FOUND | ||
| Ad4 | NOT FOUND |
| Proteins Superfamilies | Observed Intergene Distance | Expected Intergene Distance |
|---|---|---|
| Tc1<->Hc1 | 8 | 3 |
| Ne1<->Hc1 | 7 | 2 |
| Tc1<->Ad1 | 9 | 4 |
| Ne1<->Ad1 | 8 | 3 |
| Protein Superfamily | Corresponding Protein in Aclame | Aclame Phage | Seq. Identity (%) |
|---|---|---|---|
| MCP | protein:vir:99888 protein:vir:107882 protein:vir:79078 | B3 BcepMu phiE255 | 24 % 22 % 21 % |
| Portal | protein:vir:95254 protein:vir:108215 protein:vir:78161 | Felix 01 Giles Min1 | 22 % 20 % 19 % |
| TermL | protein:vir:105705 protein:vir:96782 protein:vir:78831 | ES18 phiHSIC 80alpha | 33 % 33 % 25 % |
| MTP | protein:vir:97240 protein:vir:80398 protein:vir:95133 | M6 BcepGomr PA73 | 30 % 29 % 29 % |
| Ad1 | protein:vir:94955 protein:vir:95176 protein:vir:97267 | Xp15 PA73 M6 | 25 % 20 % 19 % |
| Hc1 | protein:vir:80382 protein:vir:97237 protein:vir:104346 | BcepGomr M6 RTP | 19 % 16 % 18 % |
| Ne1 | protein:vir:95157 protein:vir:97190 protein:vir:80425 | PA73 M6 BcepGomr | 31 % 30 % 29 % |
| Tc1 | protein:vir:97211 protein:vir:80429 protein:vir:95155 | M6 BcepGomr PA73 | 34 % 28 % 28 % |