| Protein Superfamily | Index in Query Genome | Sequence Header | Detection Method |
|---|---|---|---|
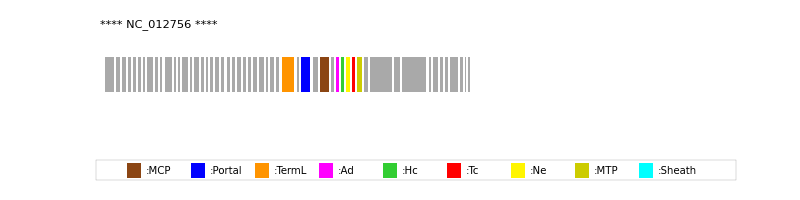
| MCP | 36 | lcl|NC_012756.1_cdsid_YP_002925169.1 | HHsearch (proba=100.00%) |
| Portal | 34 | lcl|NC_012756.1_cdsid_YP_002925167.1 | HHsearch (proba=100.00%) |
| TermL | 32 | lcl|NC_012756.1_cdsid_YP_002925165.1 | Blast (e-value=1e-173) |
| MTP | 42 | lcl|NC_012756.1_cdsid_YP_002925175.1 | Blast (e-value=5e-44) |
| Sheath | NOT FOUND | ||
| Ad1 | 38 | lcl|NC_012756.1_cdsid_YP_002925171.1 | HHsearch (proba=100.00%) |
| Hc1 | 39 | lcl|NC_012756.1_cdsid_YP_002925172.1 | HHsearch (proba=100.00%) |
| Ne1 | 40 | lcl|NC_012756.1_cdsid_YP_002925173.1 | HHsearch (proba=100.00%) |
| Tc1 | 41 | lcl|NC_012756.1_cdsid_YP_002925174.1 | HHsearch (proba=100.00%) |
| Ad2 | NOT FOUND | ||
| Hc2 | NOT FOUND | ||
| Tc2 | NOT FOUND | ||
| Ad3 | NOT FOUND | ||
| Hc3 | NOT FOUND | ||
| Ad4 | NOT FOUND |
| Protein Superfamily | Corresponding Protein in Aclame | Aclame Phage | Seq. Identity (%) |
|---|---|---|---|
| MCP | protein:vir:962 protein:vir:100172 protein:vir:100884 | bIL285 phi AT3 Lc-Nu | 37 % 29 % 29 % |
| Portal | protein:vir:80134 protein:vir:95378 protein:vir:960 | bacteriophage bv1 GBSV1 bIL285 | 53 % 51 % 47 % |
| TermL | protein:vir:959 protein:vir:80117 protein:vir:95379 | bIL285 bacteriophage bv1 GBSV1 | 53 % 50 % 50 % |
| MTP | protein:vir:968 protein:vir:80149 protein:vir:95370 | bIL285 bacteriophage bv1 GBSV1 | 45 % 40 % 40 % |
| Ad1 | protein:vir:964 protein:vir:95374 protein:vir:80144 | bIL285 GBSV1 bacteriophage bv1 | 48 % 45 % 44 % |
| Hc1 | protein:vir:95373 protein:vir:80114 protein:vir:965 | GBSV1 bacteriophage bv1 bIL285 | 36 % 35 % 29 % |
| Ne1 | protein:vir:966 protein:vir:95372 protein:vir:80116 | bIL285 GBSV1 bacteriophage bv1 | 40 % 37 % 34 % |
| Tc1 | protein:vir:80109 protein:vir:95371 protein:vir:967 | bacteriophage bv1 GBSV1 bIL285 | 35 % 32 % 32 % |