This is an old revision of the document!
Table of Contents
Protein grouping
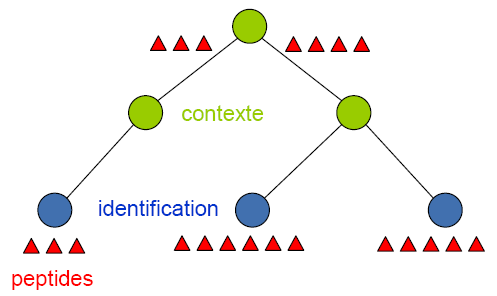
Protein grouping is done from a parent context and consist of
- creating new peptides (grouped peptide) ans matches from the union of all peptides referenced in child context (direct child if exist, otherwise more deeper childs).
- defining protein group using new set of peptides and matches associated to parent context.
Algorithm
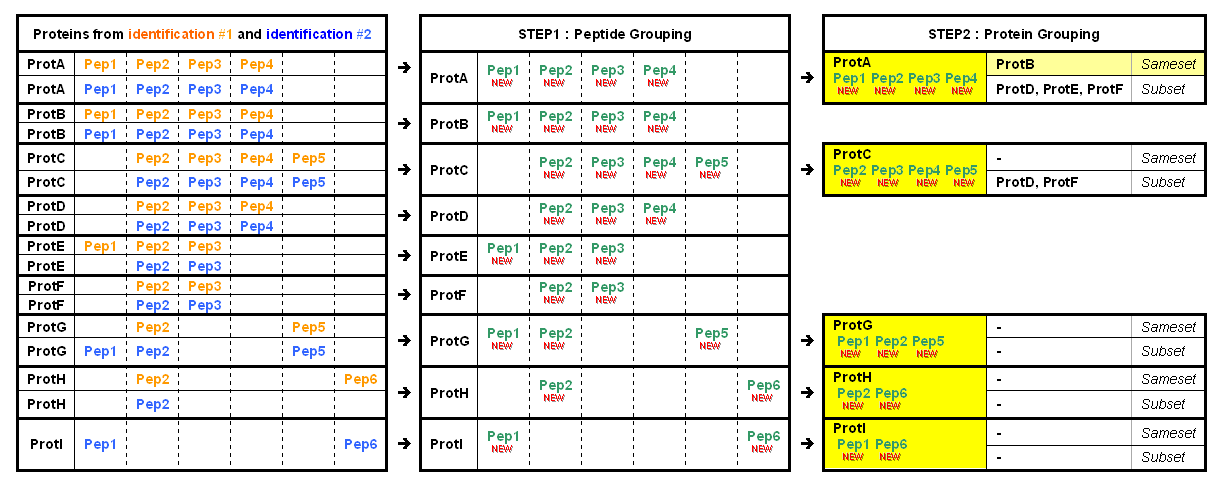
Step 1 - Peptide grouping
 Peptides from different child context or identifications - attached directly or indirectly to a context - are grouped.
Peptides from different child context or identifications - attached directly or indirectly to a context - are grouped.
Peptides must have same sequence and same experimental mass to be grouped.
Peptide grouping results in new peptides attached to the parent context and having child peptides
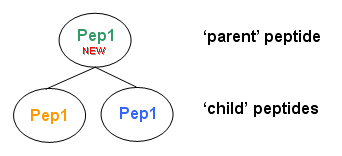 A new peptide is construct as follow :
A new peptide is construct as follow :
charge, sequence, ptm, missed cleavage and calculated massare copied from the first child peptide foundexperimental mass and retention timeare set to childs averagescoreis set to the max of child scoreschild listis set as peptides with same sequence, same mass are found- to define the
matches listassociated to new peptide, matches from all child peptides are grouped using matched protein. Created matchscoreis set to the max of all child matches scores andstart and endvalue are equal to child matches start and end.
The optional peptide filtering is applied once peptide grouping have been done and before protein grouping is run
Step 2 - Protein grouping
 Once new peptides have been created and associated to parent context, same grouping as done by Mascot® and IRMa is done. But before executing the protein grouping the list of proteins to be considered is filtered using optional protein filter. This means that proteins are filtered individually and filter is not applied to protein group level. See protein group filters page.
Once new peptides have been created and associated to parent context, same grouping as done by Mascot® and IRMa is done. But before executing the protein grouping the list of proteins to be considered is filtered using optional protein filter. This means that proteins are filtered individually and filter is not applied to protein group level. See protein group filters page.
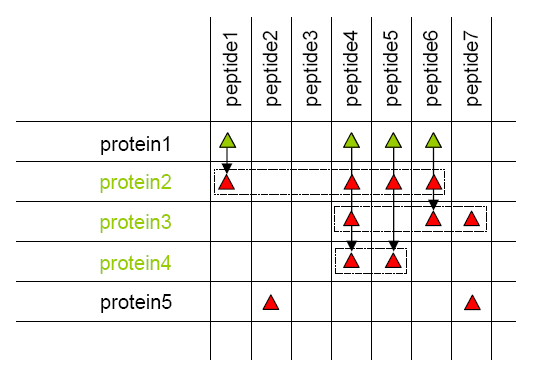
- All proteins identified by the same set of peptides are grouped together as a protein group. Proteins sharing only a sub-set of peptides are distinguished in each group. A typical protein is one of the same-set proteins. The rules used to select this typical can be specified by user.
Protein grouping results in new groups of proteins and peptides, attached to the parent context. The protein group and proteins matching properties are set as follow :
- Create a protein match for each protein of the group where the list and count of matching species is set.
- Calculate score and coverage value using all matching species.
Beware of protein grouping order
![]() You need to be carefull when grouping proteins within a tree of contexts. Let's take the following example:
You need to be carefull when grouping proteins within a tree of contexts. Let's take the following example:
Rootnode
|_ Context1
|_ F085255.dat
|_ F085256.dat
|_ F085257.dat
|_ Context2
|_ F085258.dat
|_ F085259.dat
It's possible:
- case 1 - to group proteins at the
Rootnodelevel, hEIDI will then group proteins from all the identification results, or - case 2 - to group proteins starting from the leaf contexts (
Context1andContext2), then ending with theRootnode.
At present, when launching the protein group algorithm, you can tell hEIDI to filter some proteins and/or peptides. For example, if you decide to filter proteins with a number of peptides lower than 2, it is important to understand that doing this may give different results in cases 1 & 2.
Rootnode
|_ Context1
ProtA (pep1, pep2)
|_ Context2
ProtA (pep1, pep5)
In case 2, ProtA will be filtered at an early stage (when grouping proteins in Context1 and Context2), and will not appear in the final result.
But in case 1, when grouping proteins at the Rootnode level, ProtA will 'gain' one peptide more (ProtA will be identified by 3 peptides instead of 2). So, ProtA will not be filtered and will appear in the final result.