userguide:retentiontimealignment
This is an old revision of the document!
Retention Time Alignment
Here are the steps followed by the Retention Time algorithm:
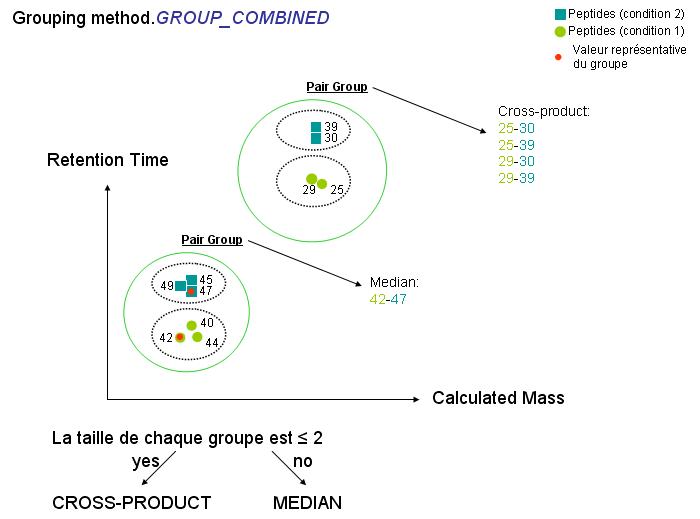
Collect species from the reference User context and predict their retention time using an external utility Collect species to align within identifications that are gathered under the reference User context Constitute pair groups between collected reference species & species to align: a pair group is created from species having same sequence & calculated mass
- Species Retention Time of the reference UserContext are predicted with NETPrediction v2.2.3378 utility using Kangas method (click here for more details). NETPrediction utility only uses species sequences to predict a Normalized Elution Time (NET) value. First, a list of 'reference' species is built with the below criteria, then the corresponding sequences are exported in order to be used by the NETPrediction utility:
- The reference species list doesn't contain any species with PTMs
- If several species exist with redundant sequences, the best score species is retained
- All the final child species, i.e. species in identifications, are collected.
a c f - tata
userguide/retentiontimealignment.1280738393.txt.gz · Last modified: 2010/08/02 10:39 by 132.168.74.230