peptidesandproteingrouping
This is an old revision of the document!
Table of Contents
Protein grouping
Algorithm
Step 1 - Peptide grouping
Peptides from different identifications - attached directly or indirectly to a context - are grouped.
Peptides must have same sequence and same experimental mass to be grouped.
Peptide grouping results in new peptides attached to the parent context and having child peptides
Step 2 - Protein grouping
Peptides from a same protein or a same protein set are grouped.
Protein grouping results in new groups of proteins and peptides, attached to the parent context.
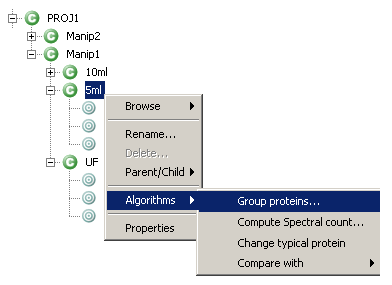
GUI - Execute protein grouping algorithm
- If needed, change settings for protein grouping. Validate.
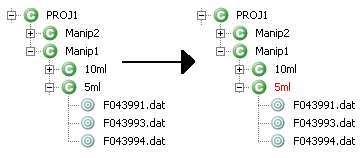
- When grouping process is finished, the color of the context name change to red in the MSIExplorer view.
Protein grouping settings:
- Peptides having same sequence and mass are gathered together, but to avoid rejecting peptides very “close” to each other, you can specify a mass tolerance (ppm)
- Optionnally, you can filter peptides according peptide length. For example, remove all peptides with a sequence lower than 6
- Optionnally, you can filter reverse proteins and/or proteins identified with a given number of low score peptides
peptidesandproteingrouping.1260948777.txt.gz · Last modified: 2009/12/16 08:32 by 132.168.74.230