how_to:proteingrouping
This is an old revision of the document!
How to group proteins within a context
Load menu
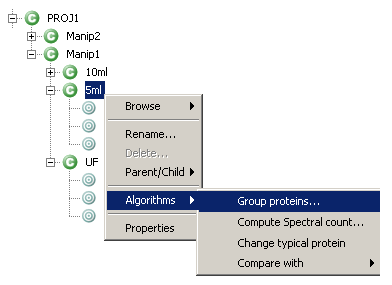
- Right-click on the context node of your choice in the MSIExplorer view to display context sensitive menu and select “Algorithms→Group proteins…”
- If needed, change settings for protein grouping (see explanations below), and
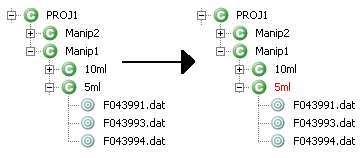
Ok - When grouping process is finished, the color of the context name change to red in the MSIExplorer view.
Parameters
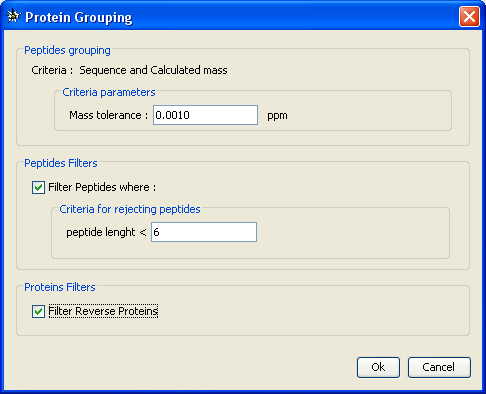
Protein grouping settings:
- Peptides having same sequence and mass are gathered together, but to avoid rejecting peptides very “close” to each other, you can specify a mass tolerance (ppm)
- Optionnally, you can filter peptides according peptide length. For example, remove all peptides with a sequence lower than 6
- Optionnally, you can filter proteins before grouping (this means filtering is done on protein and not protein groups !)
- filter reverse proteins
![]() Since hEIDI 1.11.0, you can no more filter, before grouping, proteins identified with a given number of low score peptides. This filter can be applied after grouping using Peptide count protein filter.
Since hEIDI 1.11.0, you can no more filter, before grouping, proteins identified with a given number of low score peptides. This filter can be applied after grouping using Peptide count protein filter.
how_to/proteingrouping.1311340303.txt.gz · Last modified: 2011/07/22 15:11 by 132.168.73.124