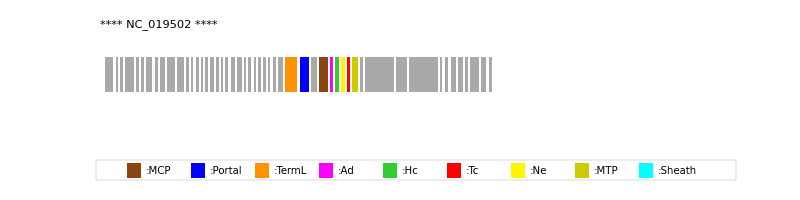
| Protein Superfamily | Index in Query Genome | Sequence Header | Detection Method |
|---|---|---|---|
| MCP | 34 | lcl|NC_019502.1_cdsid_YP_007004365.1 | HHsearch (proba=100.00%) |
| Portal | 32 | lcl|NC_019502.1_cdsid_YP_007004363.1 | HHsearch (proba=100.00%) |
| TermL | 31 | lcl|NC_019502.1_cdsid_YP_007004362.1 | Blast (e-value=0) |
| MTP | 39 | lcl|NC_019502.1_cdsid_YP_007004370.1 | Blast (e-value=2e-99) |
| Sheath | NOT FOUND | ||
| Ad1 | 35 | lcl|NC_019502.1_cdsid_YP_007004366.1 | HHsearch (proba=100.00%) |
| Hc1 | 36 | lcl|NC_019502.1_cdsid_YP_007004367.1 | HHsearch (proba=100.00%) |
| Ne1 | 37 | lcl|NC_019502.1_cdsid_YP_007004368.1 | HHsearch (proba=100.00%) |
| Tc1 | 38 | lcl|NC_019502.1_cdsid_YP_007004369.1 | HHsearch (proba=100.00%) |
| Ad2 | NOT FOUND | ||
| Hc2 | NOT FOUND | ||
| Tc2 | NOT FOUND | ||
| Ad3 | NOT FOUND | ||
| Hc3 | NOT FOUND | ||
| Ad4 | NOT FOUND |
| Protein Superfamily | Corresponding Protein in Aclame | Aclame Phage | Seq. Identity (%) |
|---|---|---|---|
| MCP | protein:vir:1268 protein:vir:102082 protein:vir:102873 | phi-105 Fah Cherry | 43 % 41 % 41 % |
| Portal | protein:vir:1266 protein:vir:102118 protein:vir:483 | phi-105 phiSM101 P27 | 40 % 34 % 33 % |
| TermL | protein:vir:81178 protein:vir:1263 protein:vir:102079 | Geobacillus virus E2 phi-105 Fah | 58 % 48 % 47 % |
| MTP | protein:vir:102037 protein:vir:102845 protein:vir:107619 | Fah Cherry Gamma | 84 % 84 % 84 % |
| Ad1 | protein:vir:81159 protein:vir:1384 protein:vir:102083 | Geobacillus virus E2 phi3626 Fah | 51 % 46 % 45 % |
| Hc1 | protein:vir:102084 protein:vir:102856 protein:vir:107606 | Fah Cherry Gamma | 86 % 86 % 86 % |
| Ne1 | protein:vir:102085 protein:vir:102875 protein:vir:107568 | Fah Cherry Gamma | 81 % 81 % 81 % |
| Tc1 | protein:vir:102086 protein:vir:102888 protein:vir:107581 | Fah Cherry Gamma | 86 % 86 % 86 % |