Table of Contents
Parse and filtering
When opening a Mascot identification file (dat file), user should specifiy parameters for Mascot parse. Indeed, the first mandatory step is to parse the dat file using free of charge parser distributed by Matrix Science.
Mascot Parse
A dialog box allow user to define parse setting. These parameters are the same as the one you can specify in Mascot Search engine. Mascot will use these informations in order to build the report.
Report Top
- Absolute : absolute number of hits to identify
- Auto : return hits with significant scores
Peptides cutoff
Ion with score less than the ion score cutoff are ignored.
Subset
Subset threshold definition: fractions core for a protein to be counted as a subset. Its score must be equal or greater than
Master_protein_score * (1-subset threshold)
If subset threshold is set to 1
*Score for a protein to be counted as a subset must be ≥ Master_protein_score * (1-1)
*Score for a protein to be counted as a subset must be ≥ Master_protein_score * (0)
*Score for a protein to be counted as a subset must be ≥ 0
All proteins sharing at least one peptide with the Master protein are counted as subsets
If subset threshold is set to 0
*Score for a protein to be counted as a subset must be ≥ Master_protein_score * (1-0)
*Score for a protein to be counted as a subset must be ≥ Master_protein_score * (1)
*Score for a protein to be counted as a subset must be ≥ Master_protein_score
No protein appears as a subset
This option can be problematical in case of further protein grouping
If subset threshold is set to 0.5
Score for a protein to be counted as a subset must be ≥ Master_protein_score * (1-0.5)
Score for a protein to be counted as a subset must be ≥ Master_protein_score * (0.5)
Score for a protein to be counted as a subset must be ≥ Master_protein_score/2
Only proteins whose score is at least equal to half of Master protein score are counted as subsets.
This option allows to limit the list of subset proteins to those which can be considered are the “more likely to be actually present in the sample”
Filtering
Master Protein Filter
By default the protein groups are represented by a master protein. This master protein is the same as the one defined by mascot. If it doesn't suit your needs you have the possibility to change the master proteins one by one in the interface OR, in one step for all the protein groups, by using the associated filter.
This filter use rules to determine which protein in the same set must be set as master. To have sufficient complexity you can composed multiple rules together.
![]() In a protein group the FIRST protein which matches the rules composition will be set as master.
In a protein group the FIRST protein which matches the rules composition will be set as master.
![]() If no protein in the protein group matches the rules composition, the old one is kept.
If no protein in the protein group matches the rules composition, the old one is kept.
![]() If the new master don't match some ambiguous peptides (because they belonging only to the old master) they will be deleted without asking (in contrary of manual master protein changing).
If the new master don't match some ambiguous peptides (because they belonging only to the old master) they will be deleted without asking (in contrary of manual master protein changing).
The rules
The rules are composed as following :
- The field on which the filter will work : Accession or Description of the protein
- The operation the filter will applied : Contains, Not contains, Begins with, Ends with
- The text to search in the given field (Case sensitive)
Example of rule :
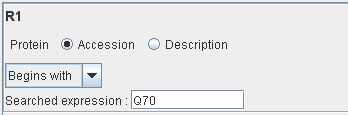
Only the proteins with their accession number beginning with “Q70” will match this rule.
Nota : you can defined up to 9 rules.
The composition
You can composed the different rules you defined to have a fine filter to set as master the proteins you want. The composition can contains :
- Rules names : R1, R2, …
- Parenthesis
- Composition operators : and, &, &&, or, |, ||, … See OGNL operator page to see all the possibilities.
As seen below the composition part of the filter window show you the translation of the composition you maked, to verify you don't do mistakes.

Examples
Example 1
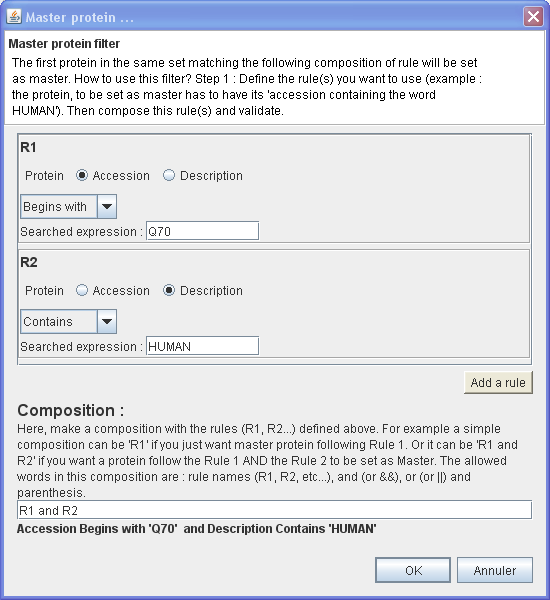
In this example, will be set as master the first protein of each protein group which have :
- Its Accession number Begins with 'Q70'
- AND
- Its Description Contains 'HUMAN'
⇒ You want to set as master only the proteins which belong to HUMAN and which have their accession number begins with 'Q70'
Example 2
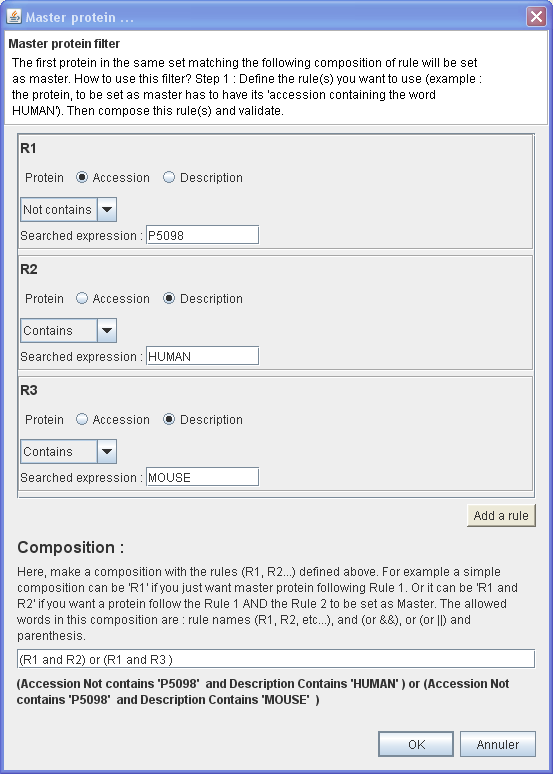
In this example, will be set as master the first protein of each protein group which have :
- Its Accession number not containing 'P5098'
- AND
- Its Description Contains 'HUMAN'
OR
- Its Accession number not containing 'P5098'
- AND
- Its Description Contains 'MOUSE'
⇒ You want to set as master only protein belong to HUMAN or MOUSE but never a particular protein which have an accession containing 'P5098'.
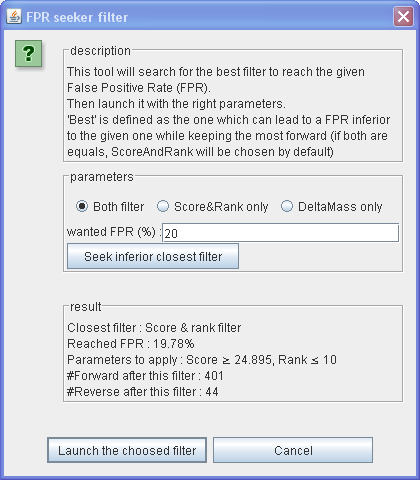
FPR Seeker Filter
The False Positive Rate (FPR) Seeker Filter will search for the best filter to reach the given FPR.
Then launch it with the right parameters.
Nota : 'Best' is defined as the one which can lead to a FPR inferior to the given one while keeping the most forward (if both are equals, ScoreAndRank will be chosen by default)
The FPR is calculated as 2xReverse/(Reverse+Forward). Where Reverse is the number of peptide matching on a Reverse protein and Forward is the number of peptide matching on a Forward protein (peptide matching on both type are not counted).
In IRMa you can check the FPR in the statistics window, into the first tab.
How to use
Result
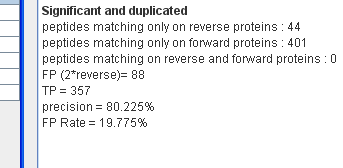
The result FPR can be seen in statistics window :
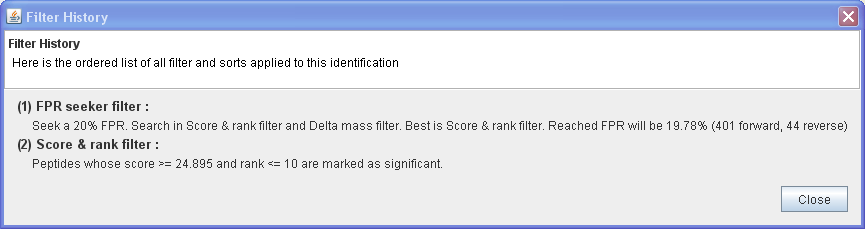
And the chained filter can be verified in Tools ⇒ Filters ⇒ Filter History:
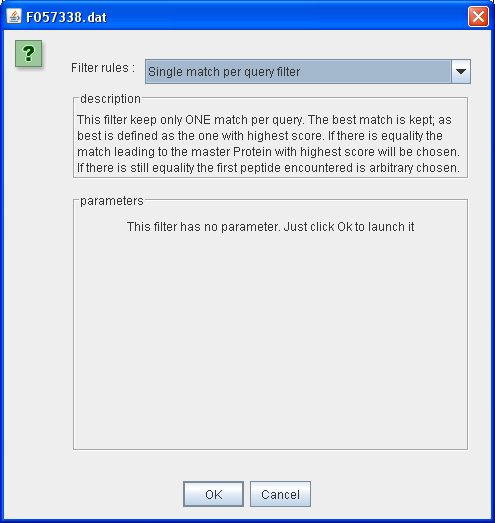
Single Match per Query Filter
This filter keep only ONE match per query. The best match is kept; as best is defined as the one with highest score. If there is equality the match leading to the master Protein with highest score will be chosen. If there is still equality the first peptide encountered is arbitrary chosen.
Example 1
In this example the query n°20 has 4 matches leading to protein (AGVLAEVR, LNIPTTR, NLLADLR, LDICQLR) (Fig 1). As the first (AGVLAEVR) have the higher score (23.15, Fig 2) all the others will be mark as ambiguous by the filter.

Fig 1
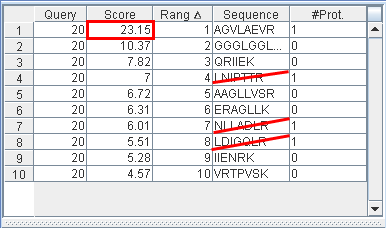
Fig 2
Example 2
The query n°1060 has 2 matches leading to proteins (LLIYGASTR & LLIYGATSR). Due to T & S swapping the two matches has the same score (35.14). As the second one (LLIYGATSR) leads to a protein with the higher score (212 opposed to 127) the first one will be mark as ambiguous.
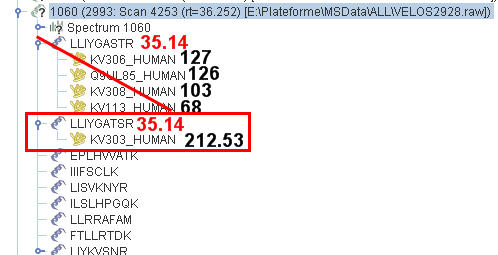
Nota : in that example, apply the Score & Rank filter with parameters to only kept rank 1 peptides would have lead to mark the second one as ambiguous, in contrary of the Single Match per Query Filter