Table of Contents
Graphical User Interface
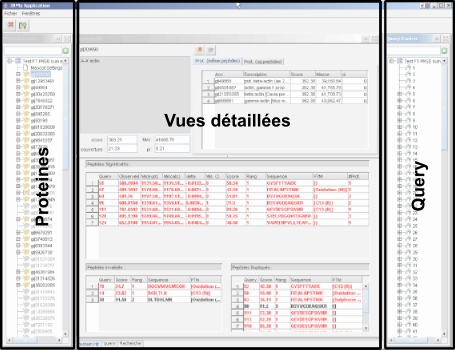
IRMa main window is splitted in three panels : the left one allow users to browse protein hits, the rigth one shows submitted queries and peptide sequences matching this queries. The center panel contains tabs representing Protein hit details, Search parameters, Query details and the list of identified proteins.
Browse protein hits
Center panel
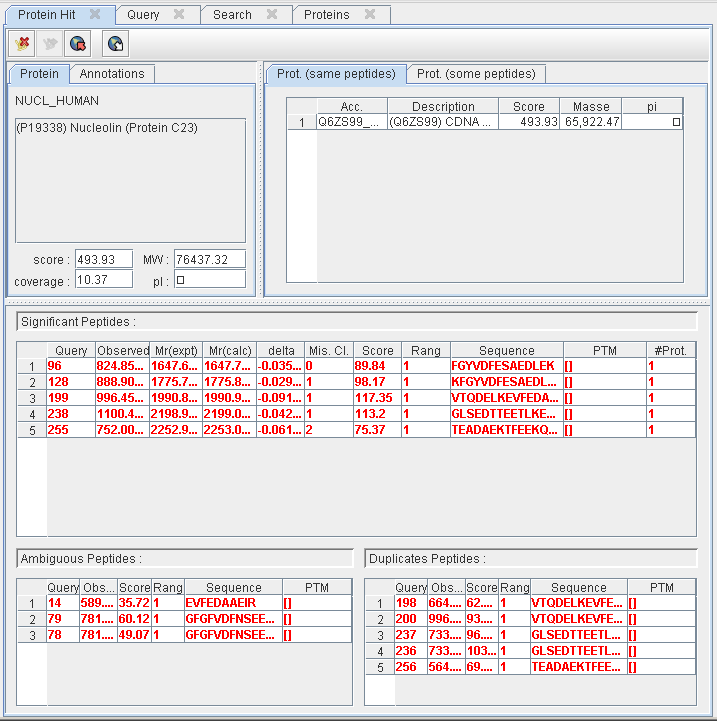
Protein Hit details
This pane displays the proteins and the set of peptides collapsed into the selected hit :
- The “main” protein details : description, score, mass, pi, coverage, etc.
- The collection of proteins matching the same set of peptides or a sub-set of this peptides.
- The set of peptides collapsed into this hit according to their category : significant, ambiguous or duplicated.
- Some additional informations associated to the “main” protein (emPAI, …)
At the top of this pane, a toolbar allow users to :
- delete the selected hit from the result if it does not contains any more significant PSM
- group the selected hit with another one if IRMa detect that the set of PSM classified as significants is a subset of PSM belonging to another hit.
The user can replace the main protein by choosing another protein among the list of proteins identified by the same set of PSM (right-clic on the protein you want to set as the new “main” protein).
By selecting a PSM, users can :
- Acces Mascot peptide view (select PSM and press space bar or rigth-clic for contextual menu).
- Classify a ambiguous a selected PSM (select PSM and press A for Ambiguous).
Query details
Displays potential peptide sequence matches of the selected query. Right-clic on a PSM to display the annotated spectra of the selected peptide and the ion fragment matches.
Search Parameters
Informations about the search : taxonomy, parameters, source file, etc.