Table of Contents
How to align retention time
See Retention Time Alignment principles if needed.
Run the Retention Time Alignment algorithm
![]() Required condition to run the Retention Time Alignment algorithm:
Required condition to run the Retention Time Alignment algorithm:
- Selected User context must be grouped
If above condition is respected, then follow these steps:
- Select the User context you want to align (You first need to group proteins for this context and save it)
- To display the settings dialog:
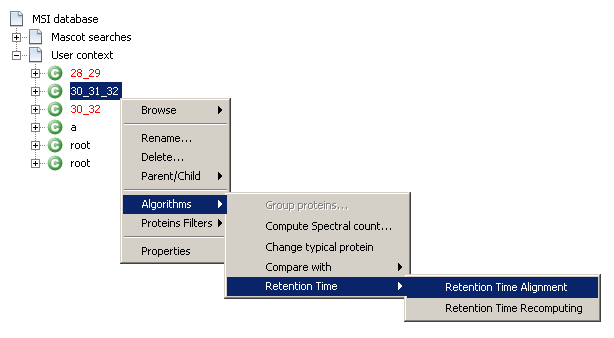
- Right-click on the selected context and select
Algorithms→Retention Time→Retention Time Alignment - Or, from the main toolbar, select
MS Identifications→Context→Algorithms→Retention Time→Retention Time Alignment
- Change algo settings options (see below) if needed and click the
Okbutton
Algo settings options (The default options are generally optimal)
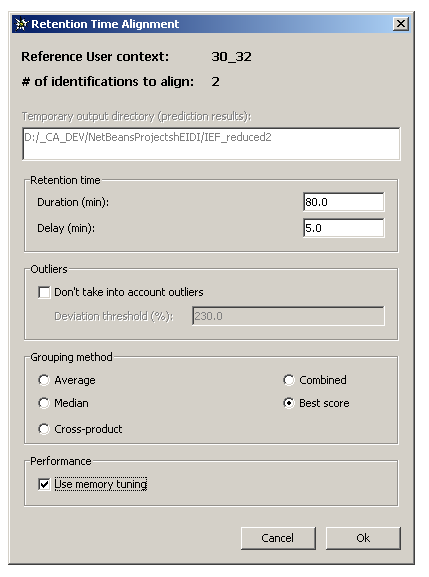
On top of the dialog, are displayed the following information:
- The reference User context
- The number of identifications to align onto the reference User context
- The directory where input/output files for NETPrediction utility are written (= the active hEIDI project directory)
Retention time panel
Duration (min): analysis time in minutesDelay (min): delay in minutes
Outliers panel
Don't take into account outliers: check this box if you want the algorithm to skip species with predicted retention time far from the others.Deviation threshold (%): if the above checkbox is selected, enter a thresh expressed in percentage. 100% represents the average absolute deviation (between RT & predicted RT). For example, a value of 200% means that species with predicted RT far about 200% from the average deviation are excluded from the calculation.
Grouping method panel
Average: average the RT values to compute the representative RT value of a species groupMedian: use median to compute the representative RT value of a species groupCross-product: use cross-product to compute the representative RT values of a species groupCombined: use median & cross-product to compute the representative RT value of a species groupBest score: use average to compute the representative RT value of a species group
Performance panel
Use memory tuning: if NOT selected, the process will be faster but memory consuming. If selected, the process will be a little bit slower but memory efficient.
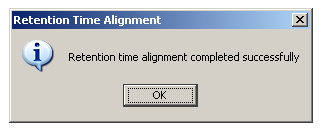
When alignment process is finished, the following dialog message appears:
View alignment results
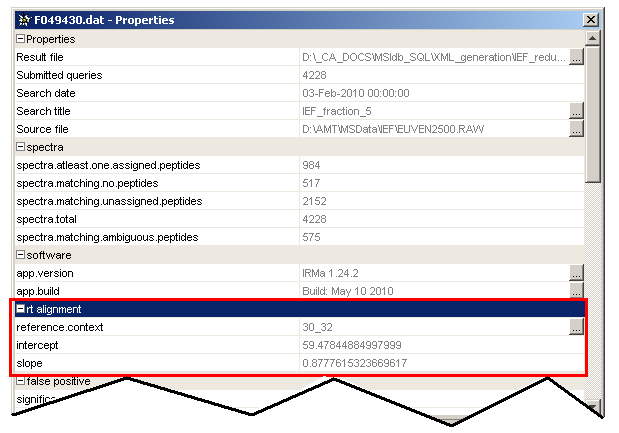
Each aligned identification has two new properties):
slope: slope of the linear regressionintercept: intercept of the linear regressionreference: reference user context
![]() You can access your identification properties either from identifications under your user context, or the under the appropriate search contexts. If you look at the identification properties just after running the Retention Time Alignment algorithm, it may not refresh immediatly.
You can access your identification properties either from identifications under your user context, or the under the appropriate search contexts. If you look at the identification properties just after running the Retention Time Alignment algorithm, it may not refresh immediatly.
Apply alignment results
To apply alignment coefficients on species, you need to Recompute Retention Time.
![]() ATTENTION
ATTENTION
During alignment process, retention time are predicted using an external utility and stored in peptide properties (peptides belonging to the reference context). These values are NOT the final recomputed retention times, they are only used by the retention time alignment algorithm.